The cover:
The kind of food an organism consumes has a broad reaching impact on its development, behavior, and lifespan. In this issue, two papers, MacNeil et al. (pp. 240–252) and Watson et al. (pp. 253–266), explore the effects of diet on these life-history traits in the nematode C. elegans. Combining nutrigenomics and network analyses, they find that different diets affect traits via distinct mechanisms. The response to diet is coupled to metabolic changes, and disruptions of some of these specific metabolic pathways correspond to inborn errors of metabolism in humans. The cover features a “native-art-inspired abstraction” of a worm eating a bacterial diet and illustrates the interconnectedness between diet, nuclear gene regulatory networks, mitochondrial networks, and their effects on life-history traits such as development and brood size.
Cover art by Lesley T. MacNeil.
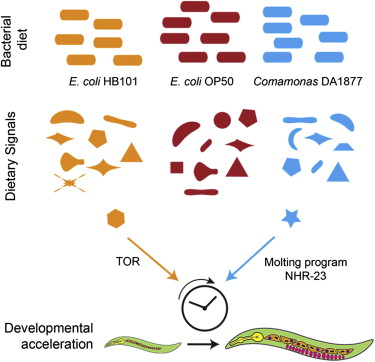
Dietary composition has major effects on physiology. Here, we show that developmental rate, reproduction, and lifespan are altered in C. elegans fed Comamonas DA1877 relative to those fed a standard E. coli OP50 diet. We identify a set of genes that change in expression in response to this diet and use the promoter of one of these (acdh-1) as a dietary sensor. Remarkably, the effects on transcription and development occur even when Comamonas DA1877 is diluted with another diet, suggesting that Comamonas DA1877 generates a signal that is sensed by the nematode. Surprisingly, the developmental effect is independent from TOR and insulin signaling. Rather, Comamonas DA1877 affects cyclic gene expression during molting, likely through the nuclear hormone receptor NHR-23. Altogether, our findings indicate that different bacteria elicit various responses via distinct mechanisms, which has implications for diseases such as obesity and the interactions between the human microbiome and intestinal cells.
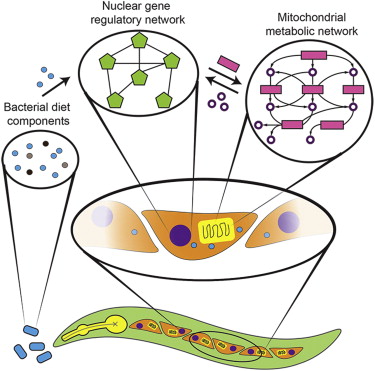
Expression profiles are tailored according to dietary input. However, the networks that control dietary responses remain largely uncharacterized. Here, we combine forward and reverse genetic screens to delineate a network of 184 genes that affect the C. elegans dietary response to Comamonas DA1877 bacteria. We find that perturbation of a mitochondrial network composed of enzymes involved in amino acid metabolism and the TCA cycle affects the dietary response. In humans, mutations in the corresponding genes cause inborn diseases of amino acid metabolism, most of which are treated by dietary intervention. We identify several transcription factors (TFs) that mediate the changes in gene expression upon metabolic network perturbations. Altogether, our findings unveil a transcriptional response system that is poised to sense dietary cues and metabolic imbalances, illustrating extensive communication between metabolic networks in the mitochondria and gene regulatory networks in the nucleus.
There’s also two free papers:
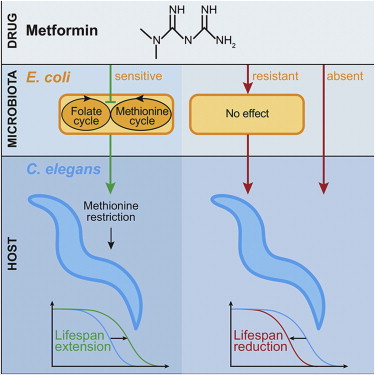
The biguanide drug metformin is widely prescribed to treat type 2 diabetes and metabolic syndrome, but its mode of action remains uncertain. Metformin also increases lifespan in Caenorhabditis elegans cocultured with Escherichia coli. This bacterium exerts complex nutritional and pathogenic effects on its nematode predator/host that impact health and aging. We report that metformin increases lifespan by altering microbial folate and methionine metabolism. Alterations in metformin-induced longevity by mutation of worm methionine synthase (metr-1) and S-adenosylmethionine synthase (sams-1) imply metformin-induced methionine restriction in the host, consistent with action of this drug as a dietary restriction mimetic. Metformin increases or decreases worm lifespan, depending on E. coli strain metformin sensitivity and glucose concentration. In mammals, the intestinal microbiome influences host metabolism, including development of metabolic disease. Thus, metformin-induced alteration of microbial metabolism could contribute to therapeutic efficacy—and also to its side effects, which include folate deficiency and gastrointestinal upset.
Systematic studies of the cancer genome have exploded in recent years. These studies have revealed scores of new cancer genes, including many in processes not previously known to be causal targets in cancer. The genes affect cell signaling, chromatin, and epigenomic regulation; RNA splicing; protein homeostasis; metabolism; and lineage maturation. Still, cancer genomics is in its infancy. Much work remains to complete the mutational catalog in primary tumors and across the natural history of cancer, to connect recurrent genomic alterations to altered pathways and acquired cellular vulnerabilities, and to use this information to guide the development and application of therapies.


Bell�articolo, molto utile! Stavo facendo le mie belle letture di post pre-nanna, dove lasciare qualche commento, con la speranza di ritorni sul mio blog, quando ho letto questo articolo! Grazie delle dritte!!!